Overview
The PL is based on a modular and extensible object-oriented architecture that allows incorporating additional functionality to the core library through external dynamically loadable modules or "plugins". In particular, the input/output system built into the PL fully relies on this plugin architecture, in such a way that the code for reading and writing data has been entirely extracted from the core of the library. This feature facilitates adding support to new file formats. The PL provides built-in routines to perform dihedral rotations, distance and derivative calculations and amino acid substitutions, among many others. The procedural API contains more than 300 base functions which can be accessed from C and C++, and from any scripting language supported by SWIG, such as Python and Perl. Plugins to calculate energy functions, simulate Monte Carlo sampling, evaluate secondary structure and solvent accessibility are already available. So far, the PL has been succesfully compiled and tested on PCs running Linux and Windows XP; however this library is not targeted at any specific computer platform or operating system.
Class hierarchy
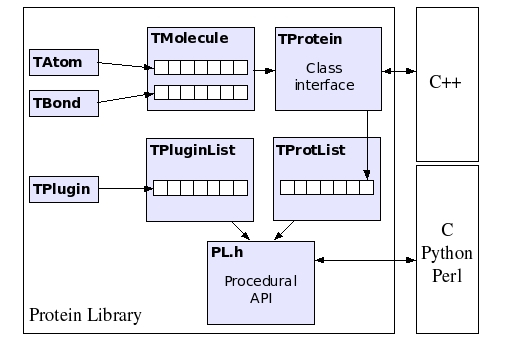
In the figure below, a simplified diagram of the class hierarchy of the PL is shown. The basic classes are TAtom and TBond, used to represent atoms and covalent bonds, respectively. There is no limitation on the size and topology of the molecules that can be stored within a TMolecule object. Its descendant, TProtein, defines the class interface of the PL, by exposing methods to calculate dihedral rotations, inter atomic distances, etc. TProtein is used in turn to construct a list of protein objects, encapsulated inside the TProtList class. The plugin subsystem is based on the TPlugin class. An arbitrary number of plugins can be loaded in TPluginList. The procedural API provides access to all the class methods of TProtein, TProtList and TPluginList, by wrapping them within standard C functions.
Plugin architecture
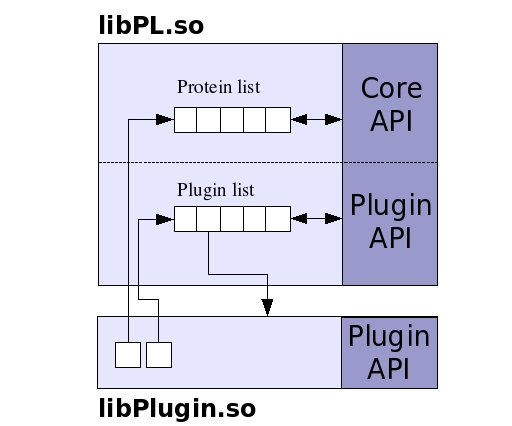
The PL supports two types of plugins: calculation plugins and input/output plugin. The calculation plugins are intended to encapsulate any computation that requires the structure information stored within the PL. These plugins can access the class interface of the PL by holding a pointer to the list of protein objects (and so they have to be written in C++). Also, a calculation plugin has access to all the other calculation plugins because it also stores a pointer to the plugin list (see next diagram). Once a plugin is loaded within the PL, it remains in memory until is explicitly unloaded. PL assigns an integer index to each one of the calculation plugins, and they are identified in the procedural API by this index.
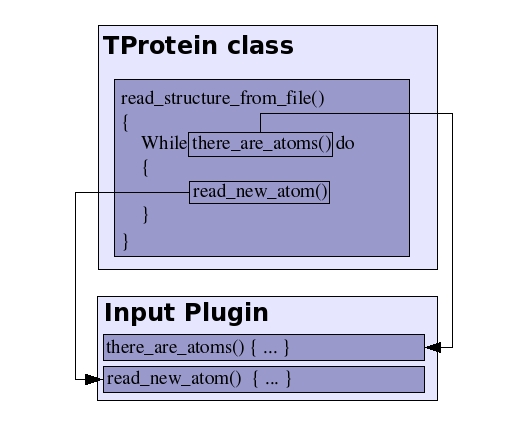
On the other hand, the IO plugins are loaded into memory only when reading/writing a structure/trajectory file. The name of the library file is passed to the member function in the TProtein object that is in charge of the reading/writing the data, together with the name of the file where the data will be read/written . The plugin is then loaded within the TProtein function, and the plugin functions are called in order to perform the elementary IO operations (reading atomic coordinates, for example). The next diagram represents the usage of the IO plugins: